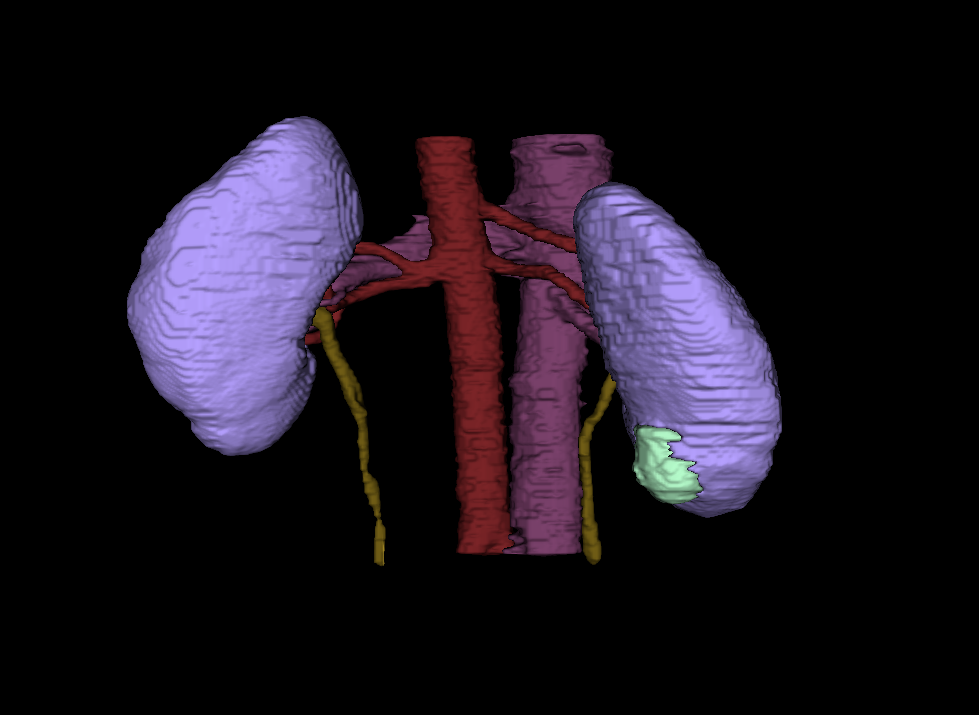
See the KiTS23 Homepage for more details, including:
- A larger dataset
- Additional contrast phases
The official repository of the 2021 Kidney and Kidney Tumor Segmentation Challenge
Current dataset version: 2.2.3 -- Official Frozen Training Set (see changelog)
- Mar 1 - Jul 1: Annotation, Release, and Refinement of Training Data (now published!)
- July 15: Further refinement of training set will be complete
- Aug 23: Deadline for Intention to Submit & Required Paper (formerly Aug 9)
- Aug 30 - Sep 13: Submissions Accepted (formerly Aug 16 - 30)
- Sep 15: Results Announced (formerly Sep 1)
- Sep 27: Satellite Event at MICCAI 2021
- July 15, 2021: The training set has been frozen!
- July 1, 2021: The training set has been released! Also we are adding a two-week buffer for final edits to be made based on community feedback, and we are pushing the challenge timeline by two weeks (see above).
- June 17, 2021: We've changed the set of classes for the challenge. See this forum post for details
- Apr 7, 2021: We've started using tags and a changelog to keep track of the dataset version
- Mar 23, 2021: A draft of the postprocessing code and some preliminary data has been merged into the master branch.
- Mar 9, 2021: A preliminary challenge homepage has been published at kits-challenge.org. You can keep tabs on the data annotation process there.
- Mar 29, 2020: A second edition of KiTS was accepted to be held in conjunction with MICCAI 2021 in Strasbourg! More information will be posted here and on the discussion forum when it becomes available.
- Install dependency for surface dice:
pip install git+https://github.com/JoHof/surface-distance.git(the original DeepMind repository is currently not working due to a missing line comment) - Clone this repository
- Install this repository by running
pip install -e .in the folder where the setup.py file is located
Start by cloning this repository, but note that the imaging is not stored here, it must be downloaded using one of the get_imaging scripts in the starter_code directory. Currently there are implementations in:
- python3:
python3 kits21/starter_code/get_imaging.py - MATLAB:
matlab kits21/starter_code/get_imaging.m - bash:
bash kits21/starter_code/get_imaging.sh - julia:
julia kits21/starter_code/get_imaging.jl
If you would like to request another implementation of get_imaging, please submit an issue.
kits21
├──data/
| ├── case_00000/
| | ├── raw/
| | ├── segmentations/
| | ├── imaging.nii.gz
| | ├── aggregated_OR_seg.nii.gz
| | ├── aggregated_AND_seg.nii.gz
| | └── aggregated_MAJ_seg.nii.gz
| ├── case_00001/
| | ├── raw/
| | ├── segmentations/
| | ├── imaging.nii.gz
| | ├── aggregated_OR_seg.nii.gz
| | ├── aggregated_AND_seg.nii.gz
| | └── aggregated_MAJ_seg.nii.gz
...
| ├── case_00299/
| | ├── raw/
| | ├── segmentations/
| | ├── imaging.nii.gz
| | ├── aggregated_OR_seg.nii.gz
| | ├── aggregated_AND_seg.nii.gz
| | └── aggregated_MAJ_seg.nii.gz
└── ├── kits.json
This is different from KiTS19 because unlike 2019, we now have multiple annotations per "instance" and multiple instances per region.
Consider the "kidney" label in a scan: most patients have two kidneys (i.e., two "instances" of kidney), and each instance was annotated by three independent people. That case's segmentations/ we would thus have
kidney_instance-1_annotation-1.nii.gzkidney_instance-1_annotation-2.nii.gzkidney_instance-1_annotation-3.nii.gzkidney_instance-2_annotation-1.nii.gzkidney_instance-2_annotation-2.nii.gzkidney_instance-2_annotation-3.nii.gz
along with similar collections for cyst, and tumor regions. The aggregated_<X>_seg.nii.gz file is a result of aggregating all of these files by various methods indicated by <X>:
- OR: A voxel-wise "or" or "union" operator
- AND: A voxel-wise "and" or "intersection" operator
- MAJ: Voxel-wise majority voting
This folder holds code snippets for viewing and manipulating the data. See Usage for more information.
This folder contains code used to process and import data from the annotation platform. As a participant, there's no reason you should need to run this code, it's only meant to serve as a reference.
This challenge will feature significantly more data, several annotations per case, and a number of additional annotated regions. The accepted proposal can be found on Zenodo, but the most up-to-date information about the challenge can be found on the KiTS21 homepage.
KiTS was first held in conjunction with MICCAI 2019 in Shenzhen. A paper describing that challenge was published in Medical Image Analysis [html] [pdf].
@article{heller2020state,
title={The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 Challenge},
author={Heller, Nicholas and Isensee, Fabian and Maier-Hein, Klaus H and Hou, Xiaoshuai and Xie, Chunmei and Li, Fengyi and Nan, Yang and Mu, Guangrui and Lin, Zhiyong and Han, Miofei and others},
journal={Medical Image Analysis},
pages={101821},
year={2020},
publisher={Elsevier}
}