-
-
Notifications
You must be signed in to change notification settings - Fork 1.1k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Polyfit performance on large datasets - Suboptimal dask task graph #5629
Comments
|
It's the same thing: |
|
Should I close this one then? |
|
Thanks for the clear example @jbusecke . I think your example is helpful enough that we should keep this open (even if the same PR will hopefully close both this and #4554) Was about to say the same @dcherian! @aulemahal what do you think about how easy it might be to change |
|
Potentially a useful comparison: here is the task graph for the same problem posed to import xarray as xr
import dask.array as dsa
def linear(x, m, b):
return m * x + b
da = xr.DataArray(dsa.random.random((4,6, 100), chunks=(1,2,100)), dims=['x','y', 'time'])
fit = da.curvefit('time', linear).curvefit_coefficients |
Should be easy! The code was written without apply_ufunc for a few reasons, one being that using |
|
You could write a wrapper that does |
|
|
|
In reading through #5933, I realized there's an alternate solution that preserves the desirable feature of fitting along chunked dimensions (at least for skipna=False) Since we are only ever fitting along one dimension, we can apply the reshape-other-dimensions operation blockwise. As long as we reshape back in a consistent manner (again blockwise) this is identical to our current stacking approach, but substantially more efficient. In particular, it avoids merging chunks. def reshape_block(arr, output_chunks):
"""
Blockwise reshape array to match output_chunks
This method applies a reshape operation blockwise, so it only makes sense if
you do not care about the order of elements (along reshaped axes) after reshaping.
The order of elements will depend on the chunking of ``arr``.
It is substantially more efficient than a usual reshape, which makes sure array elements
appear in a numpy-like order regardless of chunking.
Based on https://github.com/dask/dask/issues/4855#issuecomment-497062002
"""
from itertools import product
import numpy as np
from dask.array import Array
from dask.base import tokenize
from dask.highlevelgraph import HighLevelGraph
name = "reshape-block-" + tokenize(arr)
ichunks = tuple(range(len(chunks_v)) for chunks_v in arr.chunks)
ochunks = tuple(range(len(chunks_v)) for chunks_v in output_chunks)
dsk = {
(name, *ochunk): (np.reshape, (arr.name, *ichunk), oshape)
for ichunk, ochunk, oshape in zip(
product(*ichunks), product(*ochunks), product(*output_chunks)
)
}
graph = HighLevelGraph.from_collections(name, dsk, dependencies=[arr])
res = Array(graph, name, output_chunks, arr.dtype, meta=arr._meta)
return res
def polyfit(da, dim, skipna=False):
import itertools
import numpy as np
da = da.transpose(..., dim)
arr = da.data
do_blockwise_reshape = arr.ndim > 1 and any(
len(chunks) > 1 for chunks in arr.chunks
)
if do_blockwise_reshape:
output_chunks = (tuple(map(np.product, product(*arr.chunks[:-1]))),) + (
arr.chunks[-1],
)
other_dims = tuple(dim_ for dim_ in da.dims if dim_ != dim)
reshaped = xr.DataArray(
reshape_block(arr, output_chunks), dims=("__reshaped__", dim)
)
else:
reshaped = da
fit = reshaped.polyfit(dim, 1, skipna=skipna)
if do_blockwise_reshape:
result = xr.Dataset()
for var in fit:
result[var] = (
("degree",) + other_dims,
reshape_block(fit[var].data, ((2,),) + arr.chunks[:-1]),
)
for dim_ in other_dims:
result[dim_] = da[dim_]
result["degree"] = fit["degree"]
return result
else:
return fitHere's the graph 1 chunk along time (like @jbusecke's example, and skipna=True) And when the time dimension is chunked (and skipna=False) older versionfrom itertools import product
import numpy as np
from dask.array import Array
from dask.array.utils import assert_eq
from dask.base import tokenize
from dask.highlevelgraph import HighLevelGraph
def reshape_block(arr, output_chunks):
"""
Blockwise reshape array to match output_chunks
This method applies a reshape operation blockwise, so it only makes sense if
you do not care about the order of elements (along reshaped axes) after reshaping.
The order of elements will depend on the chunking of ``arr``.
It is substantially more efficient than a usual reshape, which makes sure array elements
appear in a numpy-like order regardless of chunking.
Based on https://github.com/dask/dask/issues/4855#issuecomment-497062002
"""
name = "reshape-block-" + tokenize(arr)
ichunks = tuple(range(len(chunks_v)) for chunks_v in arr.chunks)
ochunks = tuple(range(len(chunks_v)) for chunks_v in output_chunks)
dsk = {
(name, *ochunk): (np.reshape, (arr.name, *ichunk), oshape)
for ichunk, ochunk, oshape in zip(
product(*ichunks), product(*ochunks), product(*output_chunks)
)
}
graph = HighLevelGraph.from_collections(name, dsk, dependencies=[arr])
res = Array(graph, name, output_chunks, arr.dtype, meta=arr._meta)
return res
# assumes we're collapsing axes (0,1) and preserving axis 2
output_chunks = (tuple(map(np.product, product(*arr.chunks[:2]))),) + (arr.chunks[-1],)
reshaped = reshape_block(arr, output_chunks)
# make sure that we roundtrip
actual = reshape_block(reshaped, arr.chunks)
assert_eq(actual, arr) # Truedef new_polyfit(da):
arr = da.data
output_chunks = (tuple(map(np.product, product(*arr.chunks[:2]))),) + (
arr.chunks[-1],
)
reshaped = xr.DataArray(reshape_block(arr, output_chunks), dims=("xy", "time"))
fit = reshaped.polyfit("time", 1, skipna=True)
result = xr.Dataset()
for var in fit:
result[var] = (
("degree", "x", "y"),
reshape_block(fit[var].data, ((2,),) + arr.chunks[:-1]),
)
result["x"] = da["x"]
result["y"] = da["y"]
result["degree"] = fit["degree"]
return result
poly = da.polyfit("time", 1, skipna=False)
xr.testing.assert_allclose(poly, new_polyfit(da)) # True! |
|
@dcherian Is that something that could be implemented in apply_ufunc? Not being able to apply along a chunked dim I think happens quite often, right? |
|
The problem is that there is no general solution here. You have to write a chunk-aware function and then use |
Sorry, what case does your suggestion not handle? It would be really nice to support chunked core dims if we are going to the effort of rewriting polyfit. |
I interpreted this as asking whether this general idea could be implemented in apply-ufunc, to which the answer is "no" (I think) because it's pretty specific. At best we put this logic in a wrapper function and call that with This should handle all current use-cases of |
|
I was thinking the general idea of reshape_block seems like a clever trick to get around the apply_ufunc constraints and could be used for some other functions as well. But maybe there aren't that many functions that could make use of it. |
The idea seems similar to what we used in xhistogram, which uses blockwise to apply a function that reshapes the data in that block to be 1D. EDIT: But in that case our algorithm was one that could be applied blockwise, |
To clarify, the constraint here is that
Yes, same idea. Though when you do it that way, you have to implement the function you're applying (that xhistogram PR reimplements |
|
The explosion in chunk sizes will be fixed in the next dask release, task graph is still relatively complex though |
|
Very nice! yeah I think @jbusecke's example is the case where the "blockwise reshaping" isn't really needed because the outer most axis has chunk size 1. let's leave it open for the more general solution. |
Closes pydata#5629 1. Use Variable instead of DataArray 2. Use `reshape_blockwise` when possible following pydata#5629 (comment)
Closes pydata#5629 1. Use Variable instead of DataArray 2. Use `reshape_blockwise` when possible following pydata#5629 (comment)
Closes pydata#5629 1. Use Variable instead of DataArray 2. Use `reshape_blockwise` when possible following pydata#5629 (comment)
* Optimize polyfit Closes #5629 1. Use Variable instead of DataArray 2. Use `reshape_blockwise` when possible following #5629 (comment) * clean up little more * more clean up * Add one comment * Update doc/whats-new.rst * [pre-commit.ci] auto fixes from pre-commit.com hooks for more information, see https://pre-commit.ci * fix whats-new * Update doc/whats-new.rst Co-authored-by: Maximilian Roos <5635139+max-sixty@users.noreply.github.com> --------- Co-authored-by: pre-commit-ci[bot] <66853113+pre-commit-ci[bot]@users.noreply.github.com> Co-authored-by: Maximilian Roos <5635139+max-sixty@users.noreply.github.com>




What happened:
I am trying to calculate a linear trend over a large climate model simulation. I use rechunker to chunk the data along the horizontal dimensions and make sure that the time dimension (along which I want to calculate the fit) is not chunked.
In my realistic example, this blows up the memory of my workers.
What you expected to happen:
I expected this to work very smoothly because it should be embarassingly parallel (no information of sourrounding chunks is needed and the time dimension is complete in each chunk).
Minimal Complete Verifiable Example:
I think this minimal example shows that the task graph created is not ideal
When I apply polyfit I get this
Now the number of chunks has decreased to 4? I am not sure why, but this indicates to me that my problem might be related to #4554.
When I look at the task graph it seems that this explains why for very large dataset the computation blows up:
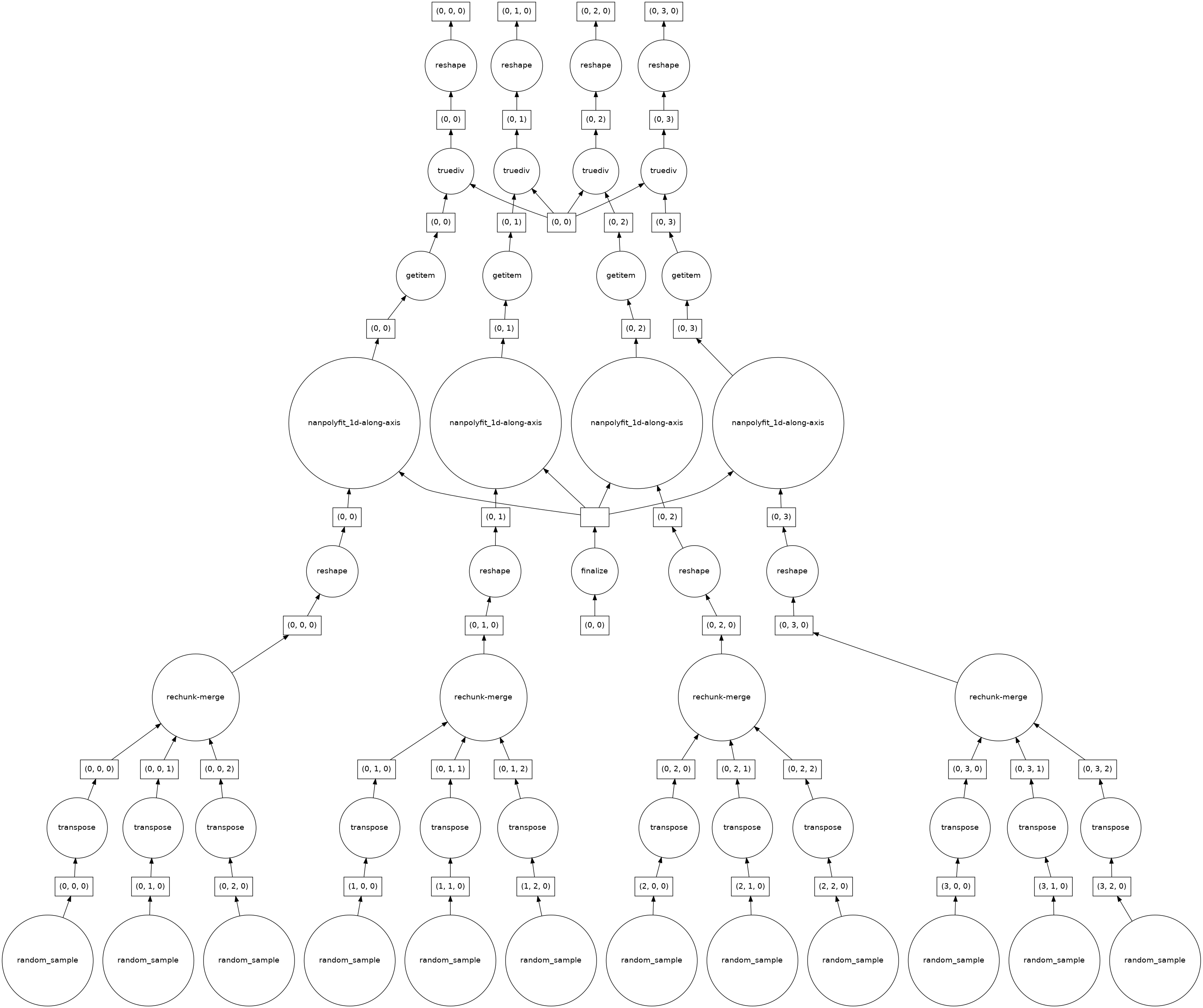
As I said before I would expect this calculation to be fully parallelizable, but there seems to be some aggregation/rechunking steps in the bottom layer. How 'bad' these get (e.g. how many input chunks get lumped together in the
rechunk/mergestep, depends on the chunking structure of the horizontal axes.cc @TomNicholas
Anything else we need to know?:
Environment:
Output of xr.show_versions()
INSTALLED VERSIONS
commit: None
python: 3.8.8 | packaged by conda-forge | (default, Feb 20 2021, 16:22:27)
[GCC 9.3.0]
python-bits: 64
OS: Linux
OS-release: 5.4.89+
machine: x86_64
processor: x86_64
byteorder: little
LC_ALL: C.UTF-8
LANG: C.UTF-8
LOCALE: en_US.UTF-8
libhdf5: 1.10.6
libnetcdf: 4.7.4
xarray: 0.17.0
pandas: 1.2.4
numpy: 1.20.2
scipy: 1.6.2
netCDF4: 1.5.6
pydap: installed
h5netcdf: 0.11.0
h5py: 3.2.1
Nio: None
zarr: 2.7.1
cftime: 1.4.1
nc_time_axis: 1.2.0
PseudoNetCDF: None
rasterio: 1.2.2
cfgrib: 0.9.9.0
iris: None
bottleneck: 1.3.2
dask: 2021.04.1
distributed: 2021.04.1
matplotlib: 3.4.1
cartopy: 0.19.0
seaborn: None
numbagg: None
pint: 0.17
setuptools: 49.6.0.post20210108
pip: 20.3.4
conda: None
pytest: None
IPython: 7.22.0
sphinx: None
The text was updated successfully, but these errors were encountered: