-
Notifications
You must be signed in to change notification settings - Fork 29
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
refactor(docs): move from reST to Markdown (#17)
Convert docs from reStructuredText to Markdown so that the changelog file is compatible with Release Please.
- Loading branch information
Showing
4 changed files
with
267 additions
and
312 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,8 @@ | ||
| # Authors | ||
|
|
||
| The list of contributors in alphabetical order: | ||
|
|
||
| - [Ana Trisovic](http://orcid.org/0000-0003-1991-0533) | ||
| - [Daniel Prelipcean](https://orcid.org/0000-0002-4855-194X) | ||
| - [Marco Donadoni](https://orcid.org/0000-0003-2922-5505) | ||
| - [Tibor Simko](https://orcid.org/0000-0001-7202-5803) |
This file was deleted.
Oops, something went wrong.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,259 @@ | ||
| # REANA example - LHCb Rare Charm Decay Search | ||
|
|
||
| [](https://forum.reana.io) | ||
| [](https://raw.githubusercontent.com/reanahub/reana-demo-lhcb-d2pimumu/master/LICENSE) | ||
|
|
||
| ## About | ||
|
|
||
| This analysis example attempts to reproduce | ||
| [LHCb rare charm decay study](https://cds.cern.ch/record/1543929) published in | ||
| [Phys. Lett. B 724 (2013) 203-212](https://www.sciencedirect.com/science/article/pii/S0370269313004747?via%3Dihub). | ||
|
|
||
|  | ||
|
|
||
| These decays are very rare, which makes their observation extremely challenging. The LHC | ||
| produces a lot of charm particles $D$, but it also produces a much greater number of | ||
| other particles which can be mistaken for the signal. It is necessary to develop an | ||
| effective strategy to identify the signal events in the large data sample. The event | ||
| selection strategy is implemented in three stages: the trigger selection, stripping | ||
| selection and the multivariate analysis. | ||
|
|
||
| In the [paper](https://cds.cern.ch/record/1543929), you can lean more about the | ||
| theoretical background and motivation to study this decay. | ||
|
|
||
| This example is based on this | ||
| [analysis-case-study](https://github.com/atrisovic/analysis-case-study). | ||
|
|
||
| ## Analysis structure | ||
|
|
||
| Making a research data analysis reproducible basically means to provide "runnable | ||
| recipes" addressing (1) where is the input data, (2) what software was used to analyse | ||
| the data, (3) which computing environments were used to run the software and (4) which | ||
| computational workflow steps were taken to run the analysis. This will permit to | ||
| instantiate the analysis on the computational cloud and run the analysis to obtain (5) | ||
| output results. | ||
|
|
||
| ### 1. Input data | ||
|
|
||
| The data being used is currently only available on request. It consists of a 10 GB ROOT | ||
| file. | ||
|
|
||
| ### 2. Analysis code | ||
|
|
||
| You can straightforwardly run the analysis step by step as explained below: | ||
|
|
||
| ```console | ||
| $ mkdir -p results/tmp && mkdir -p logs | ||
| $ root -b -q 'Optimise.C("${data}", "results")' | tee logs/optimise.log | ||
| $ root -b -q 'ModelFixing.C("${data}", "results/tmp/PhiModels.root")' | tee logs/model_fixing.log | ||
| $ root -b -q 'OSMassFit.C("${data}", "results/tmp/PhiModels.root", "results")' | tee logs/massfit.log | ||
| ``` | ||
|
|
||
| Headers and Plot style: | ||
|
|
||
| - [lhcbStyle.C](lhcbStyle.C) - Plot formats specific to the LHCb collaboration. | ||
|
|
||
| - [RooFitHeaders.h](RooFitHeaders.h) - Libraries and packages needed to perform the | ||
| fitting. | ||
|
|
||
| #### Step 1: Optimisation stage [Optimise.C](Optimise.C) | ||
|
|
||
| Optimisation is the process where combinations of different variable cuts are evaluated | ||
| in order to maximise the signal yield and reduce the background. In this analysis, the | ||
| optimisation study is performed to choose the combined BDT and particle identification | ||
| (PID) selection criteria that maximise the expected statistical significance. The result | ||
| of the script is the heat map called `2D_Optimisation_Pi.pdf`. The higher significance | ||
| (in yellow) means cleaner signal. The optimal significance is found to be at the cuts of | ||
| `BDT > 0.1` and `PIDmu > 2`. | ||
|
|
||
| ```console | ||
| $ root -b -q Optimisation.C(<data_file>, <results_directory>) | ||
| ``` | ||
|
|
||
| #### Step 2: Theoretical Model Fixing stage [ModelFixing.C](ModelFixing.C) | ||
|
|
||
| Creates the theoretical model that the fit is compared to. | ||
|
|
||
| ```console | ||
| $ root -b -q ModelFixing.C(<data_file>, <tmp_phi_models>) | ||
| ``` | ||
|
|
||
| #### Step 3: Mass Fitting stage [OSMassFit.C](OSMassFit.C) | ||
|
|
||
| The fit for the signal was modelled with the sum of Crystal Ball distributions. Each | ||
| shape consists of a Gaussian core with a power law tail on opposite sides. The background | ||
| was modelled with a 2nd order Chebyshev polynomial distribution. | ||
|
|
||
| ```console | ||
| $ root -b -q OSMassFit.C(<data_file>, <tmp_phi_models>, <results_directory>) | ||
| ``` | ||
|
|
||
| ### 3. Compute environment | ||
|
|
||
| In order to be able to rerun the analysis even several years in the future, we need to | ||
| "encapsulate the current compute environment", for example to freeze the ROOT version our | ||
| analysis is using. We shall achieve this by preparing a [Docker](https://www.docker.com/) | ||
| container image for our analysis steps. | ||
|
|
||
| Some of the analysis steps will run in a pure [ROOT](https://root.cern.ch/) analysis | ||
| environment. We can use an already existing container image, for example | ||
| [reana-env-root6](https://github.com/reanahub/reana-env-root6), for these steps. | ||
|
|
||
| ### 4. Analysis workflow | ||
|
|
||
| This analysis example consists of a simple workflow the theoretical model is generated | ||
| and used for fitting. | ||
|
|
||
| The analysis workflow consists of the above mentioned stages: | ||
|
|
||
| ```console | ||
| START | ||
| | | ||
| | D2PiMuMuOS.root | ||
| | | ||
| V | ||
| +------------------------------+ | ||
| | (1) Optimisation | | ||
| | | | ||
| | $ root Optimise.C ... | | ||
| +------------------------------+ | ||
| | | ||
| | 2D_Optimisation_Pi.pdf | ||
| | MuMuMass_Pi.pdf | ||
| | PhiModels.root | ||
| | | ||
| V | ||
| +------------------------------+ | ||
| | (2) Theoretical Model Fixing | | ||
| | | | ||
| | $ root ModelFixing.C ... | | ||
| +------------------------------+ | ||
| | | ||
| | PhiModels.root | ||
| V | ||
| +------------------------------+ | ||
| | (3) Mass Fitting | | ||
| | | | ||
| | $ root OSMassFit.C ... | | ||
| +------------------------------+ | ||
| | | ||
| | low_dimuon_signal.pdf | ||
| | high_dimuon_signal.pdf | ||
| | eta.pdf | ||
| | rho_omega.pdf | ||
| | phi.pdf | ||
| | | ||
| V | ||
| STOP | ||
| ``` | ||
|
|
||
| For example: | ||
|
|
||
| ```console | ||
| $ root -b -q 'Optimise.C("data.root", "results_directory")' | ||
| $ root -b -q 'ModelFixing.C("data.root", "phimodels.root")' | ||
| $ root -b -q 'fitdata.C("data.root", "phimodels.root", "results_directory")' | ||
| ``` | ||
|
|
||
| Note that you can also use [CWL](http://www.commonwl.org/v1.0/) or | ||
| [Yadage](https://github.com/diana-hep/yadage) workflow specifications: | ||
|
|
||
| - [workflow definition using CWL](workflow/cwl/workflow.cwl) | ||
| - [workflow definition using Yadage](workflow/yadage/workflow.yaml) | ||
|
|
||
| ### 5. Output results - Mass fit | ||
|
|
||
| The result of this analysis are the following plots in various dimuon mass ranges. We | ||
| studied the three body decay in high dimuon and low dimuon mass range, and we did not | ||
| observe any signal. | ||
|
|
||
| | Dimuon resonances | Dimuon mass range (MeV) | Plot | | ||
| | ----------------------------- | ----------------------- | ------------------------ | | ||
| | Three body decay (low dimuon) | 250 - 525 | `low_dimuon_signal.pdf` | | ||
| | $\eta$ | 525 - 565 | `eta.pdf` | | ||
| | $\rho , \omega$ | 565 - 850 | `rho_omega.pdf` | | ||
| | $\phi$ | 850 - 1250 | `phi.pdf` | | ||
| | Three body (high dimuon) | 1250 - 2000 | `high_dimuon_signal.pdf` | | ||
|
|
||
| The plots can be found in the `mass_fits` folder at the end of the execution. | ||
|
|
||
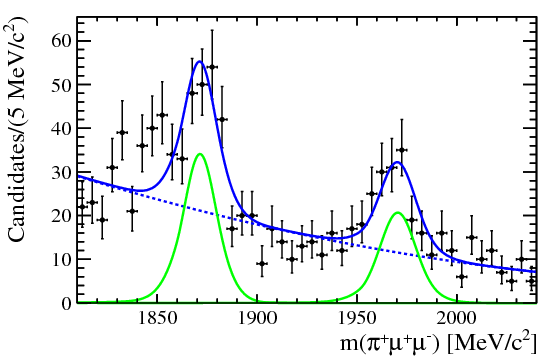
| One of the final plots, representing the $\phi$ contribution, is shown below. | ||
|
|
||
| 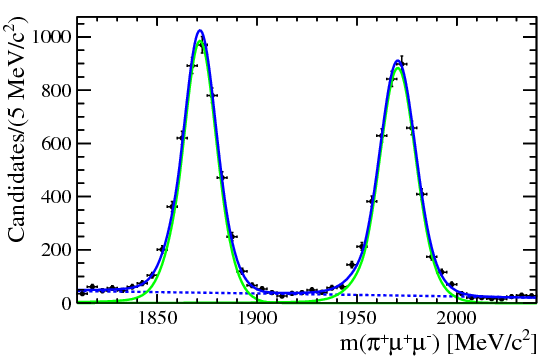 | ||
|
|
||
| 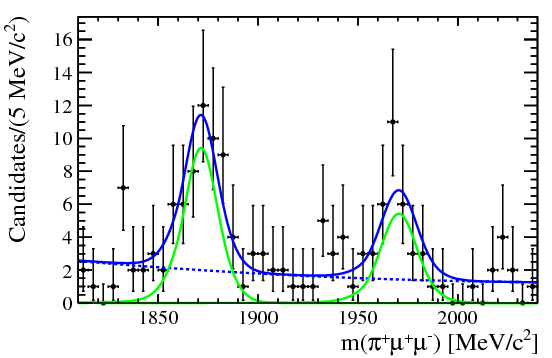 | ||
|
|
||
| 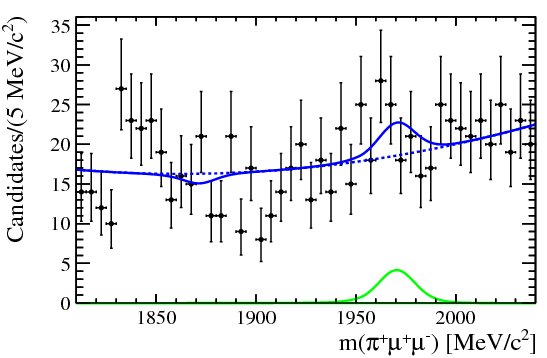 | ||
|
|
||
| 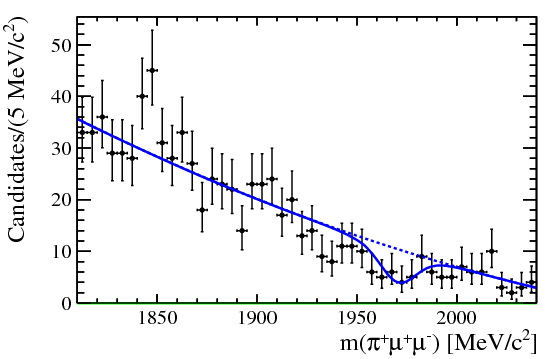 | ||
|
|
||
|  | ||
|
|
||
| ## Running the example on REANA cloud | ||
|
|
||
| We start by creating a [reana.yaml](reana.yaml) file describing the above analysis | ||
| structure with its inputs, code, runtime environment, computational workflow steps and | ||
| expected outputs: | ||
|
|
||
| ```yaml | ||
| version: 0.6.0 | ||
| inputs: | ||
| files: | ||
| - Optimise.C | ||
| - ModelFixing.C | ||
| - OSMassFit.C | ||
| - RooFitHeaders.h | ||
| - lhcbStyle.C | ||
| - data/D2PiMuMuOS.root | ||
| parameters: | ||
| data: data/D2PiMuMuOS.root | ||
| workflow: | ||
| type: serial | ||
| specification: | ||
| steps: | ||
| - environment: 'docker.io/reanahub/reana-env-root6' | ||
| commands: | ||
| - mkdir -p results/tmp && mkdir -p logs | ||
| - root -b -q 'Optimise.C("${data}", "results")' | tee logs/optimise.log | ||
| - root -b -q 'ModelFixing.C("${data}", "results/tmp/PhiModels.root")' | tee logs/model_fixing.log | ||
| - root -b -q 'OSMassFit.C("${data}", "results/tmp/PhiModels.root", "results")' | tee logs/massfit.log | ||
| outputs: | ||
| files: | ||
| - results/2D_Optimisation_Pi.pdf | ||
| - results/MuMuMass_Pi.pdf | ||
| - results/low_dimuon_signal.pdf | ||
| - results/high_dimuon_signal.pdf | ||
| - results/eta.pdf | ||
| - results/rho_omega.pdf | ||
| - results/phi.pdf | ||
| ``` | ||
| We can now install the REANA command-line client, run the analysis and download the | ||
| resulting plots: | ||
| ```console | ||
| $ # create new virtual environment | ||
| $ virtualenv ~/.virtualenvs/myreana | ||
| $ source ~/.virtualenvs/myreana/bin/activate | ||
| $ # install REANA client | ||
| $ pip install reana-client reana-cluster | ||
| $ # connect to some REANA cloud instance | ||
| $ export REANA_SERVER_URL=https://reana.cern.ch/ | ||
| $ export REANA_ACCESS_TOKEN=XXXXXXX | ||
| $ # create new workflow | ||
| $ reana-client create -n my-analysis | ||
| $ export REANA_WORKON=my-analysis | ||
| $ # upload input code and data to the workspace | ||
| $ reana-client upload ./*.C ./*.h ./data | ||
| $ # start computational workflow | ||
| $ reana-client start | ||
| $ # ... should be finished in about 15 minutes | ||
| $ reana-client status | ||
| $ # list output files | ||
| $ reana-client ls | grep ".pdf" | ||
| $ # download generated plots | ||
| $ reana-client download | ||
| ``` | ||
|
|
||
| Please see the [REANA-Client](https://reana-client.readthedocs.io/) documentation for | ||
| more detailed explanation of typical `reana-client` usage scenarios. |
Oops, something went wrong.