-
Notifications
You must be signed in to change notification settings - Fork 1
2016_incquery
The latest and last release of EMF-IncQuery is v1.1. Since March 1st, EMF-IncQuery is merged into the VIATRA project and it's new name is VIATRA Queries. You can read this blog post for more details.
In this course, we will use EMF-IncQuery v1.1 but you are free to use VIATRA Queries v1.2 in your homework by using the integration or CI updatesites. Milestone build will available at the end of April according to the plans.

homesite: https://www.eclipse.org/incquery/
Navigate to the homesite of VIATRA and search for update sites at the download page: https://eclipse.org/viatra/downloads.php
Find the old EMF-IncQuery release update site and copy it to your clipboard (don't leave white space): http://download.eclipse.org/viatra/incquery/updates/release
Switch back to your Eclipse instance and select the Help/Install New Software...
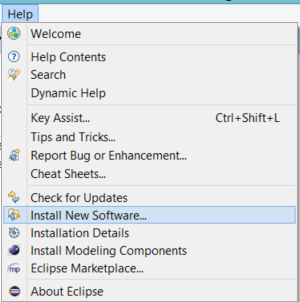
Paste the copied URL to the Work with field, than press Enter. When the view is updated, select the EMF-IncQuery SDK. Tick the Contact all update sites during install... field. Press Next, then Next and finally Finish. After the install process, you should restart Eclipse.

For a faster installation, advanced users can untick out the Contact all update sites during install... field, but they have to install Xtend and Xtext technologies manually.
-
Clone and import the following projects from this git repository: https://github.com/FTSRG/mdsd-examples
hu.bme.mit.mdsd.erdiagram hu.bme.mit.mdsd.erdiagram.edit hu.bme.mit.mdsd.erdiagram.editor -
Run as Eclipse Application. (Normally, this is not required to develop queries, but we will use the Query Explorer, which needs our erdiagram ecore model installed)
-
Import the following project to the runtime Eclipse and check the instance model.
hu.bme.mit.mdsd.erdiagram.examplediagrams hu.bme.mit.mdsd.erdiagram.example -
Create a new IncQuery project in the host Eclipse and name it to
hu.bme.mit.mdsd.erdiagram.queries. -
Create a new query definition in a package named
hu.bme.mit.mdsd.erdiagram.queriesand a file namedqueries.eiq. Also add two simple queries (and don't forget to save and build):package hu.bme.mit.mdsd.erdiagram.queries // The following imports the ecore model, // you can use auto-completion by hitting ctrl+space after the quotation mark import "hu.bme.mit.mdsd.erdiagram" pattern entity(e){ Entity(e); } pattern entityName(entity, name) { Entity.name(entity, name); }
As you can see, every pattern have a unique name and several parameters. Inside the body of the patterns, there are different constraints. Our first example describes a type constraint and the second one a feature constraint. It states that entity variable is of eClass Entity and its name attribute is the value of name variable.
Query Explorer is the primary debug tool for debugging IncQuery patterns at runtime. To open the view: Window/Show View/Others/EMF-IncQuery/Query Explorer or you can simply press the CTRL + 3 shortcut and start to type the name of the view. On the left side of the view, there will be patterns inherited from the host eclipse. The middle part will show you the matches of the patterns. To achieve this, we have to load a model into the view:
- Make sure to save and leave the focus on the opened queries.eiq file.
- Press the green arrow button on the view.
- Open our example instance model (My.erdiagram).
- Press the green arrow button on the view.

You can also filter the match set by using the panel at the right side of the query explorer.
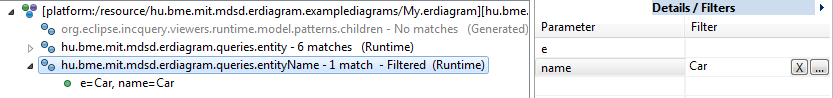
-
To get familiar with the language let's write a few validation queries, ill-formedness constraints and well-formedness constraints. Firstly, create a query to checks if the name of a
NamedElementis only an empty string:pattern emptyNamedElement(element: NamedElement) { NamedElement.name(element, ""); }
This pattern shows, that the parameters can be typed immediately in the parameters list.
-
Create a query to check if two entity has the same name:
pattern sameNamedEntities(entity1, entity2, commonName) { Entity.name(entity1, commonName); Entity.name(entity2, commonName); entity1!=entity2; }
This pattern shows the
!=(not equal) operator to select two different entites from the instance model. (Use the==operator to equality) -
Create a query to check if the name starts with a noncapital letter:
pattern entityStartsWithSmallCase(entity) { Entity.name(entity, name); check (!name.matches("^[A-Z].+")); }
This pattern shows the
checkblock where you can write a wide range of Xbase expressions (similar to Java). In this case, we define a regular expression. -
The previous queries were ill-formedness constraints. Now let's create a well-formedness constraint, which checks if an entity is well-formed.
This pattern shows how to reuse previously defined patterns as sub patterns. To do this, we use the
findkeyword then write the id of the sub pattern and finally add the variables. (Variables starting with_define don't care variables, hence you cannot use them in other lines of the pattern).This pattern also shows how to connect independent bodies in a pattern. To do this, we use the
orkeyword that states the pattern has a match if the first or the second or the third or etc body has a match.pattern badEntity(entity) { find emptyNamedElement(entity); } or { find entityStartsWithSmallCase(entity); } or { find sameNamedEntities(entity, _, _); }
A well-formed constraint ensures that there are no ill-formed structures, hence there are no matches of the
badEntity. For that, we can use thenegkey word.pattern wellFormedEntites() { neg find badEntity(_); }
-
Next, create a well-formedness constraint for
Relations, checking if it has bothRelationEndings. This will need several helper patterns too.pattern relationWithLeftEnding(r, rle) { Relation.leftEnding(r, rle); } pattern relationWithRightEnding(r, rre) { Relation.rightEnding(r, rre); } // It is required to have at least one positive constraint on a variable: pattern relationWithoutEnding(r : Relation) { neg find relationWithLeftEnding(r, _); } or { neg find relationWithRightEnding(r, _); } pattern wellFormedRelation() { N == count find relationWithoutEnding(_); N == 0; }
Notice, that using
neg findis equal to using thecountkeyword and ensure it "returns" zero. -
We can also get the number of attributes of an entity:
pattern entityAttribute(e, attr) { Entity.attributes(e, attr); } pattern attributeCount(e, N) { Entity(e); N == count find entityAttribute(e, _); }
-
Let's find the entity that is first in the alphabet:
pattern hasBiggerName(e1, e2) { Entity.name(e1, name1); Entity.name(e2, name2); check(name1 > name2); } pattern firtEntity(e : Entity) { neg find hasBiggerName(e, _); }
-
Let's find all the super entities of an entity by using transitive closure.
pattern superEntity(e,superEntity){ Entity.isA(e,superEntity); } pattern allSuperEntity(e,superEntity) { find superEntity+(e, superEntity); }
EMF-IncQuery provides facilities to create validation rules based on the pattern language of the framework. These rules can be evaluated on various EMF instance models and upon violations of constraints, markers are automatically created in the Eclipse Problems View.
The @Constraint annotation can be used to mark a pattern as a validation rule. If the framework finds at least one pattern with such annotation, a .validation project will be generated.
Annotation parameters:
- key: The parameters, which the constraint violation needs to be attached to.
-
message: The message to display when the constraint violation is found. The message may refer the parameter variables between $ symbols, or their EMF features, such as in
$Param1.name$ . - severity: "warning" or "error"
- targetEditorId: An Eclipse editor ID where the validation framework should register itself to the context menu. Use "*" as a wildcard if the constraint should be used always when validation is started.
To find a specific editor id, we can use the Plug-in Selection Spy tool with a SHIFT + ALT + F1 shortcut. Or you can just check plugin.xml in hu.bme.mit.mdsd.erdiagram.editor project.
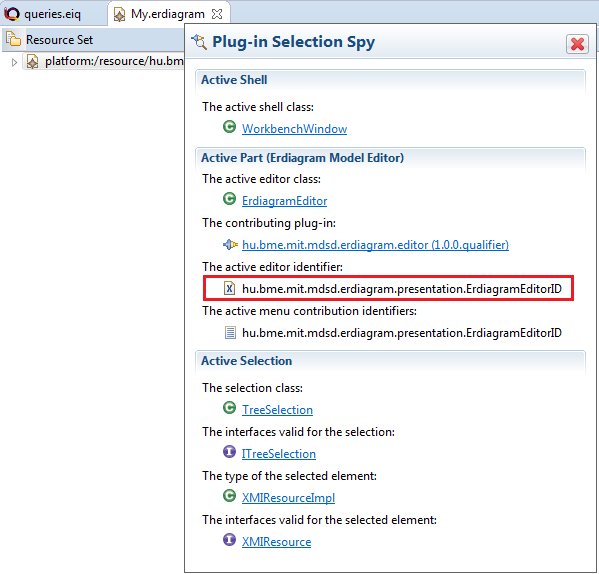
Create a constraint, using the sameNamedEntities pattern:
@Constraint(
key = {"entity1", "entity2"},
severity = "error",
message = "Two entities has the same name $commonName$",
targetEditorId = "hu.bme.mit.mdsd.erdiagram.presentation.ErdiagramEditorID"
)
pattern sameNamedEntities(entity1, entity2, commonName) {
Entity.name(entity1, commonName);
Entity.name(entity2, commonName);
entity1 != entity2;
}After build a .validation plugin project will be generated. Let's start a runtime Eclipse to install the validation plugin. After opening the model, select EMF-IncQuery Validation | Initialize EMF-IncQuery Validators on Editor. May be you have to do that twice.
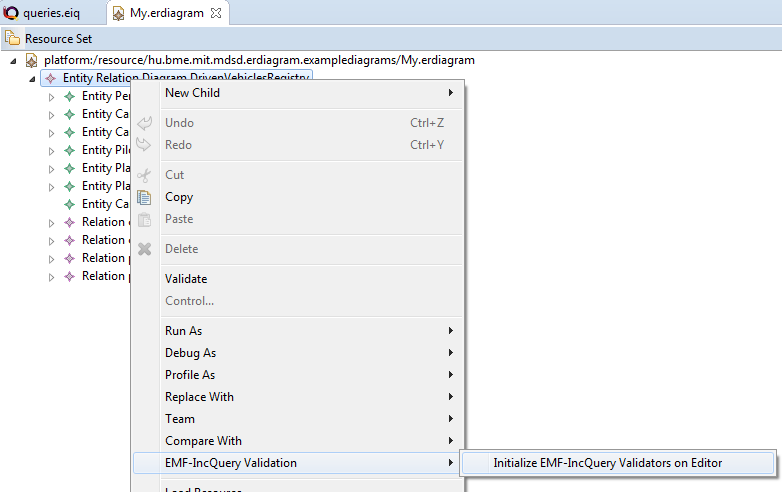
The following errors should appear (if you double click on the error, it will select the problematic EClasses):
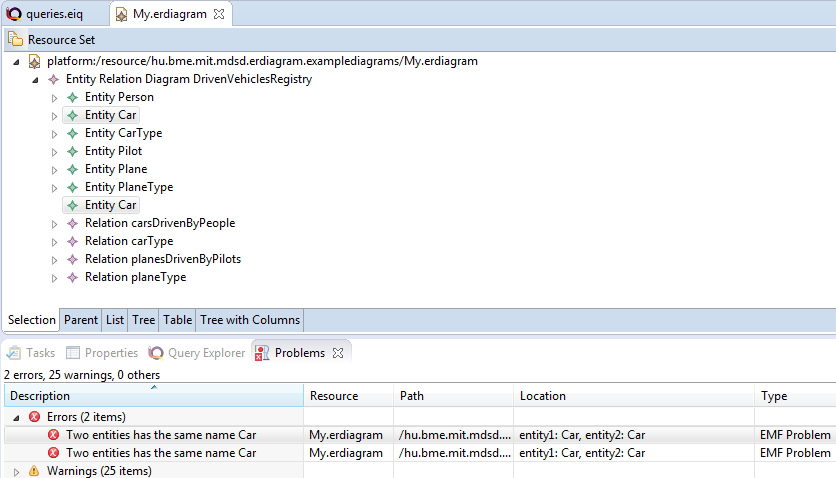
The two errors mark the same error. To solve this, add the following parameter to the constraint annotation:
symmetric = {"entity1", "entity2"}
After that, it will work as intended:

Let's create a derived feature between the RelationEndings, which returns the other RelationEnding of their parent Relation. For that the following three steps are required:
- Define a one way relation between
RelationEndings with multiplicity of [0..1] and name itotherEnding. Then set the following attributes of the feature:
- derived = true (to indicate that the value of the feature is computed from the model)
- changeable = false (to remove setter methods)
- transient = true (to avoid persisting the value into file)
- volatile = true (to remove the field declaration in the object)
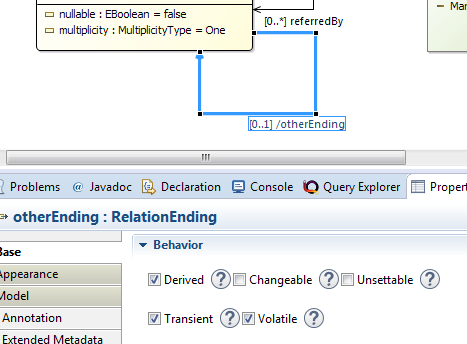
Don't forget to save the model and regenerate the model code!
- EMF-IncQuery supports the definition of efficient, incrementally maintained, well-behaving derived features in EMF by using advanced model queries and incremental evaluation for calculating the value of derived features and providing automated code generation for integrating into existing applications.
The @QueryBasedFeature annotation can be used to mark a pattern as a derived feature realization. If the framework can find out the feature from the signature of the pattern (patter name, first paramter type, second paramter type), the annotation parameters can be empty.
Annotation parameters:
- feature ="featureName" (default: pattern name) - indicates which derived feature is defined by the pattern
- source ="Src" (default: first parameter) - indicates which query parameter (using its name) is the source EObject, the inferred type of this parameter indicates which EClass generated code has to be modified
- target ="Trg" (default: second parameter) - indicates which query parameter (using its name) is the target of the derived feature
- kind ="single/many/counter/sum/iteration" (default: feature.isMany?many:single) - indicates what kind of calculation should be done on the query results to map them to derived feature values
- keepCache ="true/false" (default: true) - indicates whether a separate cache should be kept with the current value. Single and Many kind derived features can work without keeping an additional cache, as the EMF-IncQuery RETE network already keeps a cache of the current values.
Let's create the pattern, which finds the other ending and annotate it with QueryBasedFeature:
```java
@QueryBasedFeature
pattern otherEnding(ending : RelationEnding, other : RelationEnding) {
Relation.leftEnding(relation, ending);
Relation.rightEnding(relation, other);
} or {
Relation.rightEnding(relation, ending);
Relation.leftEnding(relation, other);
}
```
Save and build. IncQuery will modify the Ecore model with an annotation.
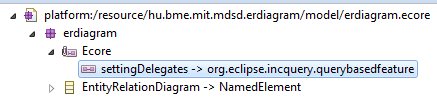
- Reload the ecore model for the genmodel and regenerate the model code. Now if you use the
relationEnding.getOtherEnding()on the model, it will return the correctRelationEnding. Note that you will need additional initialization code for IncQuery, or run it as a JUnit Plug-In test.
-
Create a pattern that detects a circle in the type hierarchy:
pattern circleInTypeHierarchy(entity) { find allSuperEntity(entity, entity); }
-
Create a pattern that detects a (transitive) diamond in the type hierarchy. Also make sure that it doesn't have more than one matches representing the same diamond:
pattern diamondInTypeHierarchy(entity1, entity2, entity3, entity4) { find allSuperEntity(entity1,entity2); find allSuperEntity(entity1,entity3); find hasBiggerName(entity2, entity3); //entity2 != entity3; find allSuperEntity(entity2,entity4); find allSuperEntity(entity3,entity4); }
-
Extend the patterns to get the inherited relations and attributes too:
pattern attribute(entity, attribute) { Entity.attributes(entity,attribute); } or { find allSuperEntity(entity, superEntity); find attribute(superEntity, attribute); }
and
pattern relation(entity1, entity2) { Relation.leftEnding.target(relation, entity1); Relation.rightEnding.target(relation, entity2); } or { find allSuperEntities(entity1, superEntity); find relation(superEntity, entity2); }
You can find the final state of the projects in this repository by checking out the IncQuery branch.
- Pattern Language: https://wiki.eclipse.org/EMFIncQuery/UserDocumentation/QueryLanguage
- Validation Framework: https://wiki.eclipse.org/EMFIncQuery/UserDocumentation/Validation
- Query Based Features: https://wiki.eclipse.org/EMFIncQuery/UserDocumentation/Query_Based_Features
- Eclipse basics
- EMF (incl. advanced topics)
- VIATRA Query
- Sirius
- Xtext+Xtend
- M2M
- Eclipse basics
- EMF (incl. advanced topics)
- VIATRA Query
- Sirius
- Xtext
- M2M
(Gradually replaced with updated content)
- Eclipse basics
- EGit
- EMF (incl. advanced topics)
- VIATRA Query
- Sirius
- Xtext
- M2M (VIATRA)
- Eclipse basics
- EGit
- EMF (incl. advanced topics)
- VIATRA Query
- Sirius
- Xtext
- M2M (VIATRA)