-
Notifications
You must be signed in to change notification settings - Fork 28
How to implement a bolus injection
Sometimes a certain amount of a chemical compound is added during the simulation of an experiment. This mean that the state variable of the respective compound changes it's value by a certain amount at a certain point of time.
Such event can be modeled by an additional input function:
INPUTS
BolusInjection C nM conc. "bolus_amount * (1/sqrt(2*pi*injection_duration^2)) * exp(-(t - injection_timepoint)^2/(2*injection_duration^2))"
The function is a scaled normal distribution (given that t indicates the independent variable) with area under the curve given by the value of bolus_amount. The injection time point is given by injection_timepoint and the duration by injection_duration.
Additionally, one has to specify a reaction that adds this input function to the respective state variable (here ProteinA):
REACTIONS
-> ProteinA CUSTOM "BolusInjection"
In the folder /Examples we provide some showcase applications Bolus_Injection_Test that can be used as a starting point for a new modeling project.
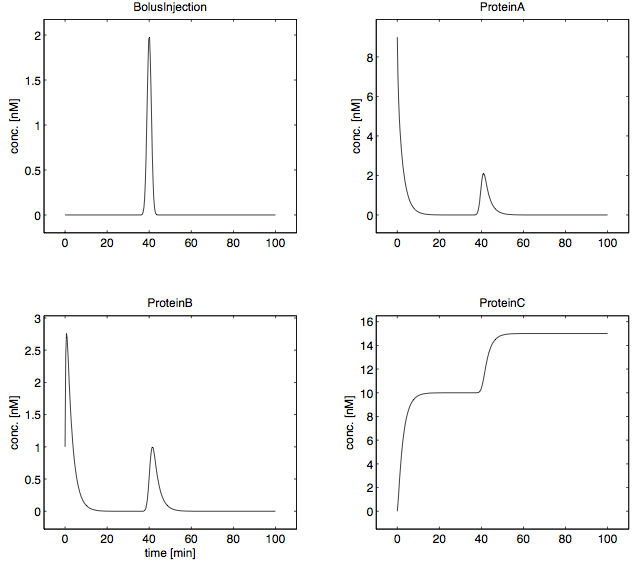
In this example a bolus of 5 nM is injected at time point 40 minutes over a period of 1 minute, as the counter state ProteinC indicates.
- Installation and system requirements
- Setting up models
- First steps
- Advanced events and pre-equilibration
- Computation of integration-based prediction bands
- How is the architecture of the code and the most important commands?
- What are the most important fields of the global variable ar?
- What are the most important functions?
- Optimization algorithms available in the d2d-framework
- Objective function, likelhood and chi-square in the d2d framework
- How to set up priors?
- How to set up steady state constraints?
- How do I restart the solver upon a step input?
- How to deal with integrator tolerances?
- How to implement a bolus injection?
- How to implement washing and an injection?
- How to implement a moment ODE model?
- How to run PLE calculations on a Cluster?