-
Notifications
You must be signed in to change notification settings - Fork 30
Import signals
Letswave 6 is able to import the following data formats :
- Biosemi BDF
- ASALAB CNT files
- Deltamed Coherence
- European Data Format (EDF, EDF+)
- Mega EEG (Neurone)
- EEGLAB SET files
- Micromed TRC files
- Brainvision VHDR files
In addition, Letswave 6 is able to import :
- data stored as ASCII text files
- data stored as a Matlab MAT variable
- data created using previous versions of Letswave
This applies to Biosemi BDF, ASALAB CNT, Deltamed Coherence, EDF, MEGA EEG, SET, TRC and VHDR files.
- Select the corresponding Import function in File > Import
Select files. Select the files you want to import.
-
For ASALAB CNT files. If the dataset contains multiple recording blocks, the option Split recording blocks in separate datasets can be used to store each block as a separate dataset.
-
For Deltamed Coherence files. In the Coherence software, you should export your data as ASCII or BIN. It is recommended to use the binary format, as this is faster and requires less space. To import the dataset, choose the text file which is created by the Coherence export function. The function will then automatically search for the associated BIN or ASCII file.
-
For MEGA EEG (Neurone software). Select the folder containing the MEGA dataset. Note that you can only import one dataset at a time. The Session number is used to define the number of the session to import.
- Select the corresponding Import function in File > Import > Import Letswave5
Select files. Select the files you want to import.
- Select the corresponding Import function in File > Import > Import Letswave 4
 Select files. Select the files you want to import. Note that these files must have been generated using the Export as structured MAT file in Letswave 4.
Select files. Select the files you want to import. Note that these files must have been generated using the Export as structured MAT file in Letswave 4.
- Select the Import from Matlab workspace function in File > Import. This function can be used to import data stored as a matrix in the Matlab workspace. The matrix can have up to four dimensions (epochs, channels, X and Y dimensions). Continuous EEG datasets will have two dimensions (channels, X). Epoched EEG datasets will have three dimensions (epochs, channels, X). Single-trial time-frequency maps would probably be stored using four dimensions (epochs, channels, X and Y).
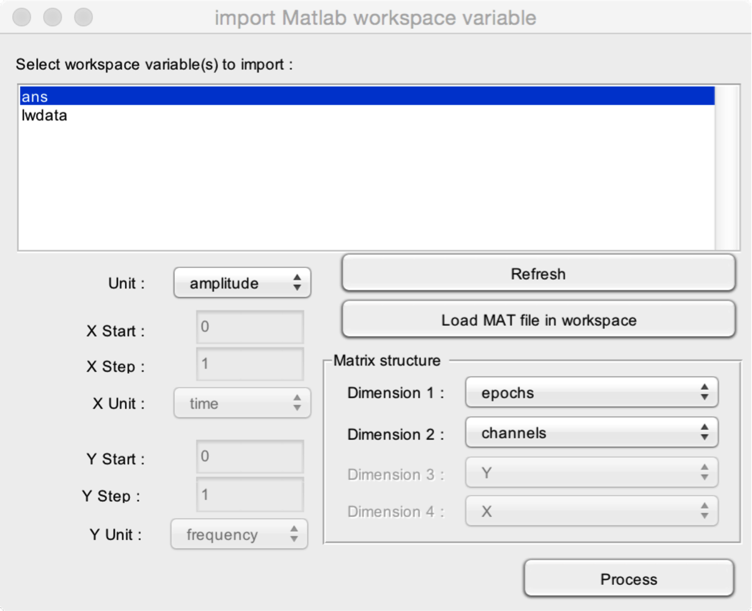
In this example, we import a workspace variable containing epoched EEG data with 10 epochs, 19 channels, and 1500 samples per epoch. Each epoch extends from -0.5 seconds to +1.0 seconds. Sampling rate is 1000 Hz. The matrix has three dimensions: epochs (10), channels (19) and X (1500). Its size is 10x19x1500.
- Select the workspace variable to import. Here, the variable to import is ans.
- Refresh to scan the content of the workspace and refreshes the list of workspace variables.
- Load MAT file in workspace to load a MAT file in the Matlab workspace.
- Define the matrix structure (i.e. how the data is organised in the matrix). In our example, the variable has three dimensions. Epochs are stored in the first dimension. Channels are stored in the second dimension. Time is stored in the third dimension (X).
- Unit defines the unit of the data stored in the variable.
- X Start defines the position of the first sample in each epoch along the X dimension. In our example, the first sample of each epoch corresponds to the signal measured at latency X=-0.5 s.
- X Step defines the interval between two consecutive samples along the X dimension. In our example, sampling rate is 1000 Hz, so X Step is 1/1000 = 0.001 s.
- X Unit defines the unit of the X dimension. In our example : X Unit = time.
- Y Start, YStep, Y Unit define the Y dimension if relevant.
- Select the corresponding Import function in File > Import > Import Letswave 4
 Select files. Select the files you want to import. Note that these files must have been generated using the Export as structured MAT file in Letswave 4.
Select files. Select the files you want to import. Note that these files must have been generated using the Export as structured MAT file in Letswave 4.
- Select the Import ASCII function in File > Import. The data stored in an ASCII (text) file must be organized as follows. Samples are stored as lines. Channels are stored as columns. The data can contain multiple epochs of equal size (i.e. equal number of lines).
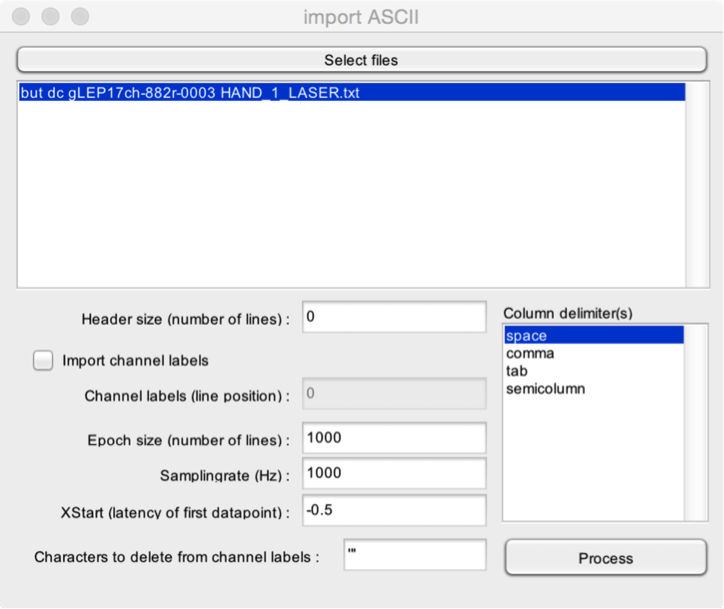
- Select files. Select one or more ASCII (text) files to import.
- Header size. Data exported as a text file often contain a header detailing the content of the text file, and providing information regarding the way the data is stored. Specify here the number of lines taken by the header.
- Import channel labels. It is possible to import channel labels, if these are stored in the text file. The channel labels must be stored on a single line, each column corresponding to a single channel label.
- Channel labels (line position). If you choose to import channel labels, you must mention the line number where channel labels are stored in the header.
- Epoch size. If the data contains several epochs, you should mention the size (number of lines) of each epoch.
- Samplingrate. The samplingrate of the data (samples/second).
- XStart. The timestamp of the first sample of each epoch (e.g. -0.5 s if the epochs extend from -0.5 to +1.0 s)
- Characters to delete from channel labels. Sometimes, bracket or hyphens are added around the channel labels. You can choose to delete these characters, for example, to recover standard International 10-5 labels.
- Column delimiters. character codes used to delimit columns in the data. It is possible to select multiple column delimiters, should that be the case.
Plugins
User interface
File
Edit
Events
- Browse and edit events
- Delete duplicate events
- Create events from level trigger
- Merge event codes and latencies
Pre-processing
- DC removal and linear detrend
- Reference
- Frequency filters
- Spatial filters (ICA)
- Epoch segmentation
- Baseline operations
- Artefact rejection and suppression
- Current source density (CSD)
- Frequency and time-frequency transforms
- Time-frequency filters
- Resample signals
- Resample signals
- Arrange signals
Post-processing
- Average
- Single-trial analysis
- Math
- Source analysis (dipole fitting)
- Find peaks in waveforms
- Global explained variance
Statistics
- Compare datasets against a constant
- Compare two datasets
- Compare more than two datasets (ANOVA)
- Compare signal amplitude at event latencies
- Bootstrap test against a reference interval
Figures