-
Notifications
You must be signed in to change notification settings - Fork 30
Spatial filters (ICA)
Blind source separation (BSS) using an Independent Component Analysis (ICA) can be used to apply a spatial filter to your data. This can be used to efficiently remove artifacts such as eye blinks and eye movements, as well as electromyographic and electrocardiographic activity (see Jung et al. 2000). In addition, ICA can be used to isolate independent sources of activity contributing to event-related potentials (see Mouraux et al. 2010).
- Compute ICA matrix
- Compute ICA matrix (merged)
- Apply ICA filter
- Assign ICA matrix
- ICA unmix signals
- ICA remix signals
Compute an Independent Component Analysis (ICA). The ICA decomposition can be performed using the runica algorithm (as implemented in EEGLAB), or using the JADER algorithm. The mixing and unmixing matrices are stored in the dataset header.
- ICA algorithm. Choose between the RUNICA algorithm or the JADER algorithm. The following methods can be used to constrain ICA:
- SQUARE (unconstrained ICA). ICA will be performed using a square matrix. The number of ICs will equal the number of channels. This is recommended when ICA is used to remove eyeblink artifacts. Note that, in some cases where the number of independent sources is far lower than the number of channels, this can lead to overfitting and the creation of spurious ICs. In these cases, it is recommended to reduce the number of ICs (see also Beckmann & Smith 2004).
- USER-DEFINED constrained ICA. ICA will be performed using a rectangular matrix. The number of ICs is defined by the user.
- PROBABILISTIC-ICA. Several methods are implemented to automatically estimate the number of independent sources in the data, and to perform ICA using this estimate. This two-step method is referred to as Probabilistic ICA (PICA). Available methods are Laplacian Estimate (LAP), Bayesian Information Criterion (BIC), Rajan & Rayner (RRN), AIC and MDL (Beckmann & Smith 2004). Note that in our experience, good results have been obtained using the RRN approach.
- Percentage of PICA estimate. If you are using PROBABILISTIC ICA, it is possible to increase or decrease the number of estimated ICs, expressed as a percentage of the estimate provided by the algorithm. This is useful to check that the obtained results are not critically dependent on the number of estimated ICs.
This function is identical to the COMPUTE ICA MATRIX function except for the fact that ICA is performed after merging multiple datasets. The obtained ICA matrix is then assigned to each original dataset. This is useful if you wish to obtain a single matrix for multiple datasets that have all been obtained during the same recording. Note that this is valid if and only if it is reasonable to assume that all the datasets can be explained by a single set of independent components, each projecting identically to the different channels across datasets.
Interactive GUI to remove independent components, e.g. independent components capturing artifacts such as eye blinks or muscle activity. Note that this function requires computing the ICA first (see Compute ICA matrix).
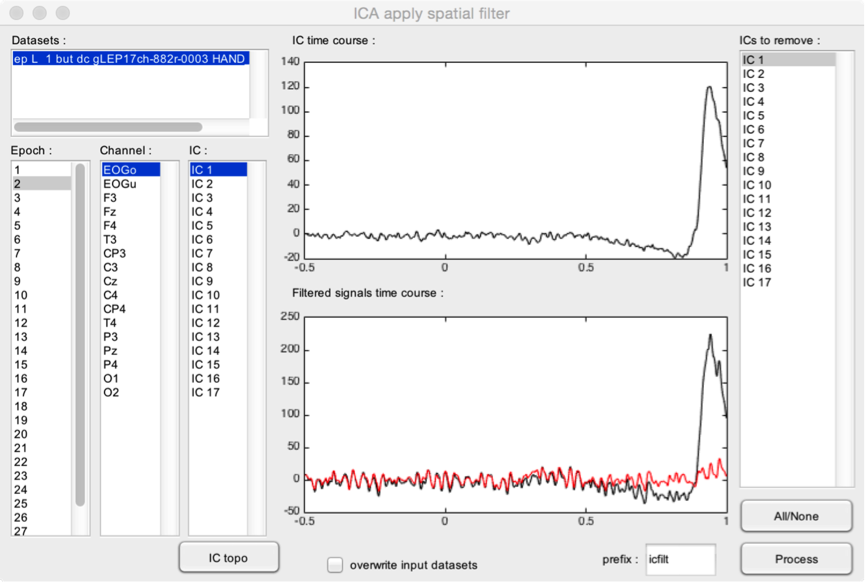
- Datasets. It is possible to process several datasets at the same time, as long as they have the same associated ICA matrix.
The upper graph shows the IC time course for the selected dataset and epoch. Use the IC listbox to change the ICs shown in this graph. In this example, we can see that IC1 captures eye movement artifacts, as evidenced by the large eye blink artifact occurring approximately 0.8 s after stimulus onset.
The lower graph shows the EEG time course for the selected dataset and epoch. The black waveform corresponds to the unfiltered signal. The red waveform corresponds to the filtered signal (i.e. the signal obtained after removing the ICs selected in the ICs to remove listbox). In this example, you can see that removing IC1 efficiently removes the eye blink artifact. You can change the displayed channel and epoch using the Channel and Epoch listbox.
- IC topo. Plot the scalp topography of the selected ICs.
- All/None. Select or unselect all ICs in the ICs to remove listbox.
Assign an existing ICA matrix of a given dataset to another dataset.
- Load dataset with ICA matrix. Select a dataset which already has an associated ICA matrix.
Unmix original signals into a set of independent components (ICs). This requires a dataset with an associated ICA matrix.
Remix independent components (ICs) into mixed signals.
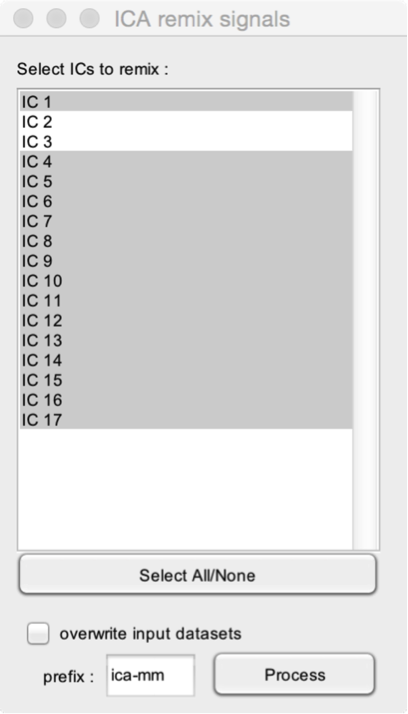
- Select ICs to remix. It is possible to remix only a selection of independent components (ICs), for example, to only keep ICs which do not contain an artifact. In this example, the remixed signals will include the signals contained in all ICs except IC2 and IC3.
Plugins
User interface
File
Edit
Events
- Browse and edit events
- Delete duplicate events
- Create events from level trigger
- Merge event codes and latencies
Pre-processing
- DC removal and linear detrend
- Reference
- Frequency filters
- Spatial filters (ICA)
- Epoch segmentation
- Baseline operations
- Artefact rejection and suppression
- Current source density (CSD)
- Frequency and time-frequency transforms
- Time-frequency filters
- Resample signals
- Resample signals
- Arrange signals
Post-processing
- Average
- Single-trial analysis
- Math
- Source analysis (dipole fitting)
- Find peaks in waveforms
- Global explained variance
Statistics
- Compare datasets against a constant
- Compare two datasets
- Compare more than two datasets (ANOVA)
- Compare signal amplitude at event latencies
- Bootstrap test against a reference interval
Figures